My new *BEAST (starbeast) species tree runs are converging wonderfully in BEAST v1.8.3, so I am off to a great start this morning. Here, I’m including a screenshot of the results of one of these runs, which I ran for 200 million generations using 3 normally distributed calibration points, as visualized in Tracer. Note the good posterior trace, very high ESS scores for all parameter estimates, and good use of burn-in (default 10% value works quite well in this case).
This is the most beautiful thing I’ve seen in my research today, or even this week!
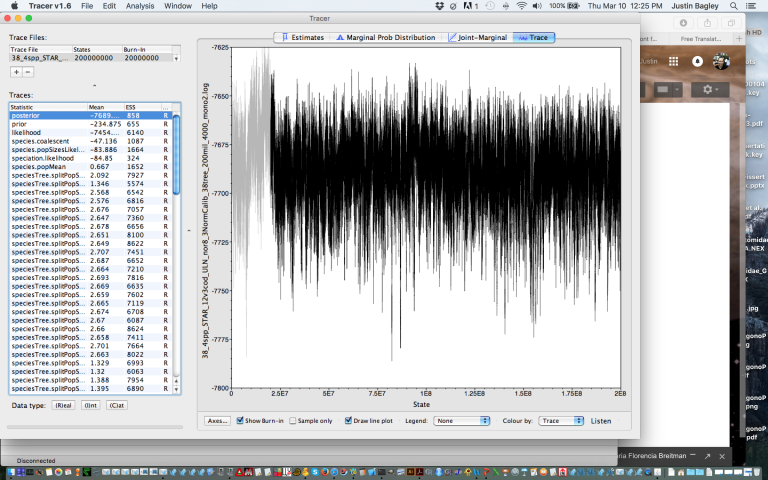
I am now checking and processing the results from multiple such runs using different species schemes (trait mapping schemes/files). They seem like they are going to give me a nice estimate of the species tree for this dataset.
Currently, the second steps of these runs are still running, using path sampling/stepping-stone sampling to calculate marginal likelihood estimates (MLE) for each model. When this is completed, I will calculate Bayes factors for each model and use them for Bayes factor species delimitation (BFD) sensu Grummer et al. (2014).
Is anyone else working on BFD analyses like this right now? Would love to hear from you.
~J
References
Grummer JA, Bryson RW Jr., Reeder TW. 2014. Species delimitation using Bayes factors: simulations and application to the Sceloporus scalaris species group (Squamata: Phrynosomatidae). Systematic Biology 63(2): 119-133.